© LuckyStep – stock.adobe.com
News • Gene-snipping bacterial defenses
CRISPR: a promising tool to fight antibiotic resistance, but...
Experts are looking to use the Nobel winning technology of Clustered regularly interspaced short palindromic repeats (CRISPR) to target resistance genes and make bacteria sensitive to first line antibiotics again; but the bacteria have ways to fight back.
In a new research review on this subject presented at the ESCMID Global Congress (formerly ECCMID) in Barcelona, Assistant Prof. Ibrahim Bitar, Department of Microbiology, Faculty of Medicine and University Hospital in Plzen, Charles University in Prague, Plzen, Czech Republic, gave an overview of the molecular biology of CRISPR technology in explaining how it can used to tackle antimicrobial resistance.
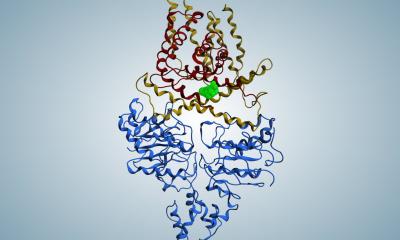
Clustered regularly interspaced short palindromic repeats (CRISPRs) and CRISPR associated genes (cas) are widespread in the genome of many bacteria and are a defence mechanism against foreign invaders such as plasmids and viruses. The CRISPR arrays are composed of a repeated array of short sequences, each originating from and exactly matching a nucleic acid sequence that once invaded the host.
Recommended article

News • For the development of CRISPR/Cas9
Nobel Prize in Chemistry goes to Emmanuelle Charpentier and Jennifer A. Doudna
The Royal Swedish Academy of Sciences has decided to award the Nobel Prize in Chemistry 2020 to Emmanuelle Charpentier from the Max Planck Unit for the Science of Pathogens, Berlin, Germany, and Jennifer A. Doudna from the University of California, Berkeley, USA, “for the development of a method for genome editing”, more commonly known as the 'gene scissors' CRISPR/Cas9.
Accompanying CRISPR sequences, there are 4-10 CRISPR-associated genes (cas), which are highly conserved and encode the Cas proteins. Cas proteins conduct adaptive immunity in prokaryotes (bacteria) based on immunological memories stored in the CRISPR array. The CRISPR/Cas system integrates a small piece of foreign DNA from invaders such as plasmids and viruses into their direct repeat sequences and will recognise and degrade the same external DNA elements during future invasions.
As the CRISPR/Cas systems integrate DNA from invading pathogens in chronical order, genotyping can be used to trace the clonality and the origin of the isolates and define them as a population of strains that were subjected to the same environmental conditions including geographic location (region) and community/hospital settings and eventually further extended to track pathogenic bacteria around human society.
The bacterial pathways are always complicated and such systems are always heavily regulated by multiple pathways. These regulated pathways must be studied in depth in order to avoid selective pressure favoring anti-CRISPR systems activation
Ibrahim Bitar
CRISPR/Cas systems can also be employed for developing antimicrobial agents: introduction of self-targeting crRNAs will effectively and selectively kill target bacterial populations. Due to the shortage of available effective antimicrobial agents in treating multidrug-resistant (MDR) infections, researchers started to search for alternative methods to fight MDR infections rather than going through the process of developing new antimicrobial agents which can go on for decades. As a result, the concept of CRISPR/Cas-based selective antimicrobials was first developed and demonstrated in 2014. Vectors coding Cas9 and guide RNAs targeting genomic loci of a specific bacterial strain/species can be delivered to the target strain via bacteriophages or conjugative bacterial strains.
In theory, delivery of the engineered CRISPR/Cas systems specifically eliminates target strains from the bacterial population, yet it is not that simple. While these systems can seem a target for manipulation/intervention, all bacteria are regulated by multiple pathways to ensure the bacteria retains control over the process. Therefore, there remain several major challenges in using this system as an antimicrobial agent.
Most methods require delivery of the re-sensitised system by conjugation; the vector is carried by a non-virulent lab strain bacteria that is supposed to go and share the vector/plasmid through conjugation. The conjugation process is a natural process that the bacteria do which results in sharing plasmids among each other (even with other species). The percentage of conjugated (successfully delivered) bacteria in the total bacterial population is critical to the re-sensitised efficiency. This process is governed by several complicated pathways.
Recommended article

Article • Bacterial defense mechanism
Antibiotic resistance: a global threat to healthcare
Antimicrobial resistance (AMR) is becoming more prevalent around the world, constituting a serious threat to public health. When bacteria acquire resistance against antibiotics, common medical procedures – for example, in surgery – become impossible due to the high infection risk. Keep reading to find out about AMR research, development of new antibiotics and antibiotic alternatives.
Bacteria also possess built-in anti-CRISPR systems, that can repair any damage caused by CRISPR-Cas systems. Defence systems that the bacteria uses to protect itself from foreign DNA often co-localise within defence islands (genomic segments that contain genes with similar function in protecting the host from invaders) in bacterial genomes; for example: acr (a gene that acts, with other similar variants, as a repressor of plasmid conjugative systems) often cluster with antagonists of other host defence functions (e.g., anti-restriction modification systems) and experts hypothesise that MGEs (mobile genetic elements) organise their counter defence strategies in “anti-defence” islands.
Assistant Professor Bitar concludes: “In summary, this method seems very promising as an alternative way of fighting antimicrobial resistance. The method uses the concept of re-sensitising the bacteria in order to make use of already available antibiotics – in other words, removing their resistance and making them vulnerable again to first-line antibiotics. Nevertheless, the bacterial pathways are always complicated and such systems are always heavily regulated by multiple pathways. These regulated pathways must be studied in depth in order to avoid selective pressure favoring anti-CRISPR systems activation, hence prevalence of resistance in a more aggressive manner.”
Source: European Society of Clinical Microbiology and Infectious Diseases
30.04.2024